Selected Publications
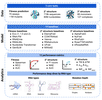
RNAGym: Large-scale Benchmarks for RNA Fitness and Structure Prediction
Preprint. Large-scale benchmarks to assess models for RNA fitness & structure prediction.
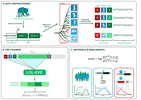
Evolutionary-Scale Enzymology Enables Biochemical Constant Prediction Across a Multi-Peaked Catalytic Landscape
Science, 2025. A microfluidic platform to measure catalytic constants for hundreds of Adenylate Kinase variants, enabling mapping of the sequence-catalysis landscape and development of superior predictive models.
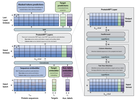
Protriever: End-to-End Differentiable Protein Homology Search for Fitness Prediction
ICML, 2025. an end-to-end differentiable framework that learns to retrieve relevant protein homologs while simultaneously training for downstream tasks, achieving state-of-the-art fitness prediction performance while being orders of magnitude faster than traditional MSA-based approaches.
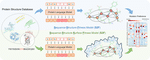
Large-scale discovery, analysis, and design of protein energy landscapes
Preprint. A multiplexed experimental approach to analyze conformational fluctuations across thousands of protein domains, revealing hidden variations that affect protein cooperativity and function.
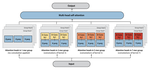
Multi-Scale Representation Learning for Protein Fitness Prediction
NeurIPS, 2024. A multimodal framework that integrates protein sequence, structure, and surface topology features to achieve state-of-the-art fitness prediction.
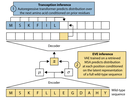
Predicting Promoter Variant Effects from Evolutionary Sequences
Preprint. A conditional autoregressive transformer model trained on 14.6 million mammalian promoter sequences that achieves state-of-the-art performance in predicting the effects of indels in human promoter regions.
Machine Learning for Functional Protein Design
Nature Biotech, 2024. A unifying framework that classifies models on the basis of their use of three core data modalities: sequences, structures and functional labels is introduced to make sense of the exploding diversity of machine learning approaches.
ProteinGym: Large-Scale Benchmarks for Protein Fitness Prediction and Design
NeurIPS, 2023. Large-scale benchmarks to assess models for protein fitness prediction and design.
ProteinNPT: Improving Protein Property Prediction and Design with Non-Parametric Transformers
NeurIPS, 2023. A conditional semi-supervised pseudo-generative model for fitness prediction and design.
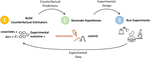
DiscoBAX: Discovery of Optimal Intervention Sets in Genomic Experiment Design
ICML, 2023. A sample-efficient method for discovering optimal sets that are both diverse and optimize the function of interest.
TranceptEVE: Combining Family-specific and Family-agnostic Models of Protein Sequences for Improved Fitness Prediction
NeurIPS, LMRL, 2022. A hybrid family-specific and family-agnostic model to achieve SOTA performance on protein fitness prediction and human variant annotation.
Tranception: Protein Fitness Prediction with Autoregressive Transformers and Inference-time Retrieval
ICML, 2022. A suite of autoregressive transformers with biological priors, augmented with inference-time retrieval, to achieve SOTA performance on protein fitness prediction.
Disease Variant Prediction with Deep Generative Models of Evolutionary Data
Nature, 2021. Deep Generative Models (Bayesian VAEs) of evolutionary sequences to predict the effects of missense mutations in human proteins.
GeneDisco: A Benchmark for Experimental Design in Drug Discovery
ICLR, 2022. A benchmark suite for evaluating active learning algorithms for experimental design in drug discovery.
Improving Black-box Optimization in VAE Latent Space Using Decoder Uncertainty
NeurIPS, 2021. A framework that uses the epistemic uncertainty of the decoder of a VAE to guide the optimization of properties of high-dimensional structured objects (e.g., molecules) in latent space.
Improving Compute Efficacy Frontiers with SliceOut
Preprint. A memory-efficient dropout-inspired scheme to train large neural networks faster with no loss in accuracy.