DiscoBAX: Discovery of Optimal Intervention Sets in Genomic Experiment Design
 DiscoBAX
DiscoBAX
Abstract
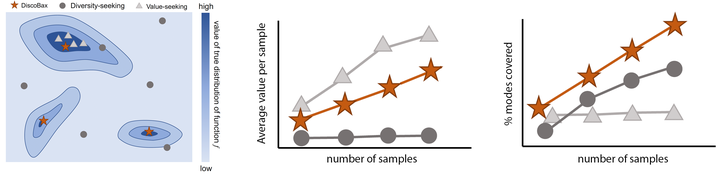
The discovery of therapeutics to treat genetically-driven pathologies relies on identifying genes involved in the underlying disease mechanisms. Existing approaches search over the billions of potential interventions to maximize the expected influence on the target phenotype. However, to reduce the risk of failure in future stages of trials, practical experiment design aims to find a set of interventions that maximally change a target phenotype via diverse mechanisms. We propose DiscoBAX, a sample-efficient method for maximizing the rate of significant discoveries per experiment while simultaneously probing for a wide range of diverse mechanisms during a genomic experiment campaign. We provide theoretical guarantees of approximate optimality under standard assumptions, and conduct a comprehensive experimental evaluation covering both synthetic as well as real-world experimental design tasks. DiscoBAX outperforms existing state-of-the-art methods for experimental design, selecting effective and diverse perturbations in biological systems.